- Vaudel et al. Proteomics 2011;11(5):996-9.
- If you use SearchGUI as part of a publication, please include this reference.
 |
v2.0.11 - Windows | ReleaseNotes |
 |
v2.0.11 - Mac and Linux | ReleaseNotes |
 |
 |
 |
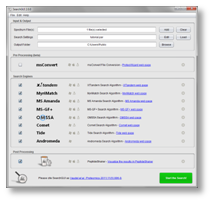
(Click on figure to see the full size version)
SearchGUI is a user-friendly open-source graphical user interface for configuring and running proteomics identification search engines, currently supporting X!Tandem, MS-GF+, MS Amanda, MyriMatch, Comet, Tide, Andromeda and OMSSA.
To start using SearchGUI, unzip the downloaded file, and double-click the SearchGUI-X.Y.Z.jar file. No additional installation required!
To visualize and analyze the search results we recommend PeptideShaker.
For developer access to the search results we recommend the use of compomics-utilities.
- From the Command Line
- Database Help
- User Defined Modifications
- Converting Spectrum Data
- Result Analysis
To start identifying peptides and proteins using SearchGUI, download the latest version, unzip the downloaded file, and double-click on the SearchGUI-X.Y.Z.jar file.
The main purpose of SearchGUI is to make it simpler to use multiple search engines at the same time. We believe that a graphical user interface would be the best choice for most users, and therefore made SearchGUI with a graphical user interface. However, it can sometimes be easier to perform a search from the command line. For example when incorporating the search into some sort of pipeline. With this in mind we have therefore included the option of using SearchGUI as a command line tool.
For details about the command line see: SearchCLI.
It is straightforward to add/edit modifications via the graphical user interface. Modifications will be available in other instances of SearchGUI and PeptideShaker for the same user/computer. Not all modifications are correctly handled by the search engines. For example, X!Tandem is not compatible with modifications at termini on specific amino acids. Using such a modification will result in nonsense matches which can be filtered out afterwards. This functionality is available by default in PeptideShaker.
SearchGUI currently only supports mgf files as the input format for the spectra. To convert your raw data to mgf we recommend using msconvert from ProteoWizard.
To visualize and analyze the SearchGUI results we recommend the use of PeptideShaker. PeptideShaker is a search engine independent platform for visualization of peptide and protein identification results from multiple search engines.
-
Does Not Start I - Do you have Java installed? Download the latest version of Java here and try again. (You only need the JRE version (and not the JDK version) to run SearchGUI.)
-
Does Not Start II - Have you unzipped the zip file? You need to unzip the file before double clicking the jar file. If you get the message "A Java Exception has occurred", you are most likely trying to run SearchGUI from within the zip file. Unzip the file and try again.
-
Does Not Start III - Is SearchGUI installed in a path containing special characters, i.e.
[,%, æ, ø, å, etc? If so, move the whole folder to a different location or rename the folder(s) causing the problem and try again. (Note that on Linux SearchGUI has to be run from a path not containing spaces). -
Unidentified Developer - If you run SearchGUI on a Mac you can get the warning "SearchGUI" can't be opened because it is from an unidentified developer. To escape this warning control-click on the file icon and then select "Open." This will give you the option of opening it regardless of its unidentified source. This only has to be done once for each SearchGUI version.
-
Search Engine Issues - Important: If you have problems with the search engines, please verify that the search engines are working outside of SearchGUI first. To test your installation run the search engine executable on the command line. This should result in output describing what the script does. If you get this, it works, and SearchGUI should run without problems. If not, see below.
-
X!Tandem XML Syntax Error - If X!Tandem gives the error "Syntax error parsing XML", the problems is most likely that the path to your database or mgf files contains special characters not supported on your operating system. If this happens try renaming the folders containing the special characters or move the files to folders not containing special characters.
-
OMSSA on Windows - Windows users might encounter issues starting OMSSA, due to missing libraries. Do solve this do the following: First, apply the following [update](http://www.microsoft.com/downloads/details.aspx?FamilyID#200b2fd9-ae1a-4a14-984d-389c36f85647&displaylang#en http://www.microsoft.com/downloads/details.aspx?FamilyID#200b2fd9-ae1a-4a14-984d-389c36f85647&displaylang#en), then apply this [security patch](http://www.microsoft.com/downloads/details.aspx?familyid#766a6af7-ec73-40ff-b072-9112bab119c2&displaylang#en http://www.microsoft.com/downloads/details.aspx?familyid#766a6af7-ec73-40ff-b072-9112bab119c2&displaylang#en).
-
Linux Support - Users wanting to use SearchGUI on Linux may have to install the search engines first, see the tools web pages for available search engine versions. Important: Please verify that the search engines are working outside of SearchGUI before using them inside SearchGUI.
-
Linux Support II - If you get problems running makeblastdb (need to prepare FASTA files for OMSSA searches) make sure that you have the required 32 bit libraries. To install the libraries you can use "sudo apt-get install ia32-libs".
-
MS Amanda on Linux - Running MS Amanda on Linux requires that you have Mono installed. Mono 3.2.1 or newer is required and the libmono-system-core4.0-cil has to be installed. To check your Mono version run "mono -V". Mono can be installed as follows: "sudo apt-get install mono-runtime" (and if needed "sudo apt-get install libmono-system-core4.0-cil" and "sudo apt-get -f"). For some Linux distributions Mono version 3.X is not directly available, you might be able to use: "sudo add-apt-repository ppa:directhex/monoxide", "sudo apt-get update" and "sudo apt-get dist-upgrade". For more help on installing Mono please see http://www.mono-project.com/download.
-
MS Amanda on Mac - Running MS Amanda on Linux requires that you have Mono installed. Mono 3.10.0 or newer is required. For help on installing Mono please see http://www.mono-project.com/download.
-
MyriMatch on Linux - If you get the error "
locale::facet::_S_create_c_locale name not valid", this can be fixed by running the command "export LC_ALL#C" before running SearchGUI/MyriMatch. To make this fix permanent, put the export line in your .bash_profile (~/.bash_profile). If you still have problems, please contact the MyriMatch developers. -
Linux and Mac OSX File Permissions - On Linux and Mac OSX you may have to edit the permissions for the executable files in order for SearchGUI to work. Allow execution for all users.
-
32 bits vs 64 bits - Please make sure that your using versions of the search engines that are compatible with your OS. Note that the latest releases of OMSSA are only available in 64 bits versions. For older versions of OMSSA see the OMSSA archive. However, it is always recommended to use the latest version if possible. Also note that OMSSA versions up until version 2.1.9 does not support precursor mass tolerance in ppm.
-
Problem Not Solved? Or Problem Not List? - Contact the developers of SearchGUI by setting up an issue describing the problem. If the issue is related to the installation of the search engines, please contact the search engine developers directly.